Mr Xiaowei Jiang (PhD Student)

Contact Details:
Email: jiangx@tcd.iePhone: +353 (0) 1 8962873
A computational approach to decipher the molecular immune response to pathogenic bacteria
A large number of new protein families emerged during the evolution of the immune response system in mammals. The great molecular differences between proteins belonging to different families involved in the response to bacterial invasions has been considered as the main cause for the recognition of pathogenic bacteria and the neutralisation of their infectious cycle. On the other hand, bacteria also present specific genetic strategies which enable them to escape from the immune response and successfully infect the host.
Despite the huge effort made to unveil the main bacterial proteins responsible for their infectivity, conclusions have been always obscured by the lack of a general view of how the interaction between bacterial and host proteins coevolves. Most of the studies have focused on the analysis of specific proteins previously reported to be responsible for the bacterial infectivity. The study of the coordinated function of proteins belonging to different families has been hampered by the lack of theoretical and experimental approaches on protein families involved on pathogenesis and their interaction with host immune proteins. Over 600 bacterial genomes have been obtained during the last few years. Meanwhile, there are 2 complete mammalian genomes to date, however there are 36 (45.57%) in assembly and 41 in progress. The amount of information available at the molecular and genome levels and the lack of rigorous studies on the analysis of the interaction of the immune system of mammals with bacterial protein families have fuelled the design of this project.
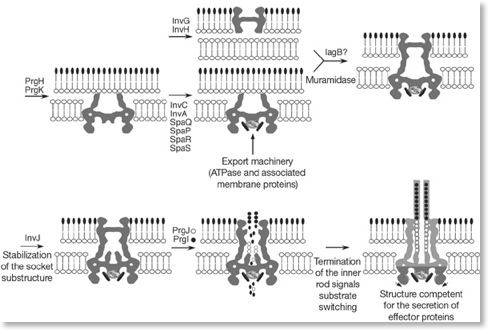
The type III secretion system (T3SS) has a central component the needle complex, which has a significant role in bacterial pathogenesis. Some bacterial pathogens use T3SS to deliver their own proteins into host cells, which can help them initiate their intra-cellular life that is pathogenic for their hosts. The above figure is a model of the assembly pathway of the needle complex by Jorge E. Galán and Hans Wolf-Watz (Nature 2006). The nomenclature of relevant proteins is from S. typhimurium.
The objectives in this project are:
- Identify patterns in proteins that contribute to the pathogenesis of bacterial pathogens.
- Identify patterns in proteins from mammalian immune response that contribute to the neutralization of bacterial pathogens.
- Investigate the relationships between those counterpart proteins and look at individual proteins in terms of molecular evolution and co-evolution.
- Explore new statistical and mathematical models for such analysis.
The results obtained in this research would be of unprecedented importance for the incoming biomedical studies and would make possible future interactions between academia and industry built on the basis of testing the proposed proteins as drug targets.
Education
- MSc (Statistics), Katholieke Universiteit Leuven, Belgium, 2007
- MSc (Bioinformatics), Katholieke Universiteit Leuven, Belgium, 2005
- BE (Computer Science and Technology), Shenyang Institute of Aeronautical Engineering, China, 2004
Publications
1. Jiang X, Fares M.A. (2009). Identifying coevolutionary patterns in Human Leukocyte Antigen (HLA) molecules. Evolution. (Epub ahead of print).





